Scientific Data Pipelines Framework - DataJoint
Data Stewards Roundtable
2026-01-27
The Problem: Scientific Data Pipelines
Problem
Raw Data
Raw Data
Process\nStep 1
Process
Step 1
Raw Data->Process\nStep 1
Process\nStep 2
Process
Step 2
Process\nStep 1->Process\nStep 2
Process\nStep 3
Process
Step 3
Process\nStep 2->Process\nStep 3
Analysis
Analysis
Process\nStep 3->Analysis
The Problem: Challenges
Manual dependency tracking Hard to reproduce resultsLost provenance informationDifficult to scale workflowsError-prone manual steps
The Problem: Typical Workflow
# Step 1: Check if raw data exists if not os.path.exists('raw_data.h5' ):raise Error("Missing raw data" )# Step 2: Check if step1 processed if not os.path.exists('step1_result.npy' ):# Step 3: Check if step2 processed if not os.path.exists('step2_result.npy' ):# But did step1 run? # Step 4: Manual tracking # Which version? Which parameters?
What is DataJoint?
Database schema = Executable workflow specification
Image source: docs.datajoint.com
Relational Workflow Model
Tables = Workflow stepsForeign keys = DependenciesSchema = Executable specification
Table Tiers
Tiers
Manual
Manual
#Lookup
#Lookup
Manual->#Lookup
_Imported
_Imported
#Lookup->_Imported
__Computed
__Computed
_Imported->__Computed
Table Structure
class Subject(dj.Manual):= """ subject_id: int --- subject_name: varchar(100) species: varchar(50) """
Manual Tables
class Session(dj.Manual):= """ -> Subject # Foreign key session_id: int # Primary key --- session_date: date # Secondary attributes notes: varchar(500) """
Lookup Tables
class #StimulusType(dj.Lookup): = """ stimulus_type: varchar(20) --- description: varchar(200) """ = ['visual' , 'Visual stimulus' ),'auditory' , 'Auditory stimulus' )
Imported Tables
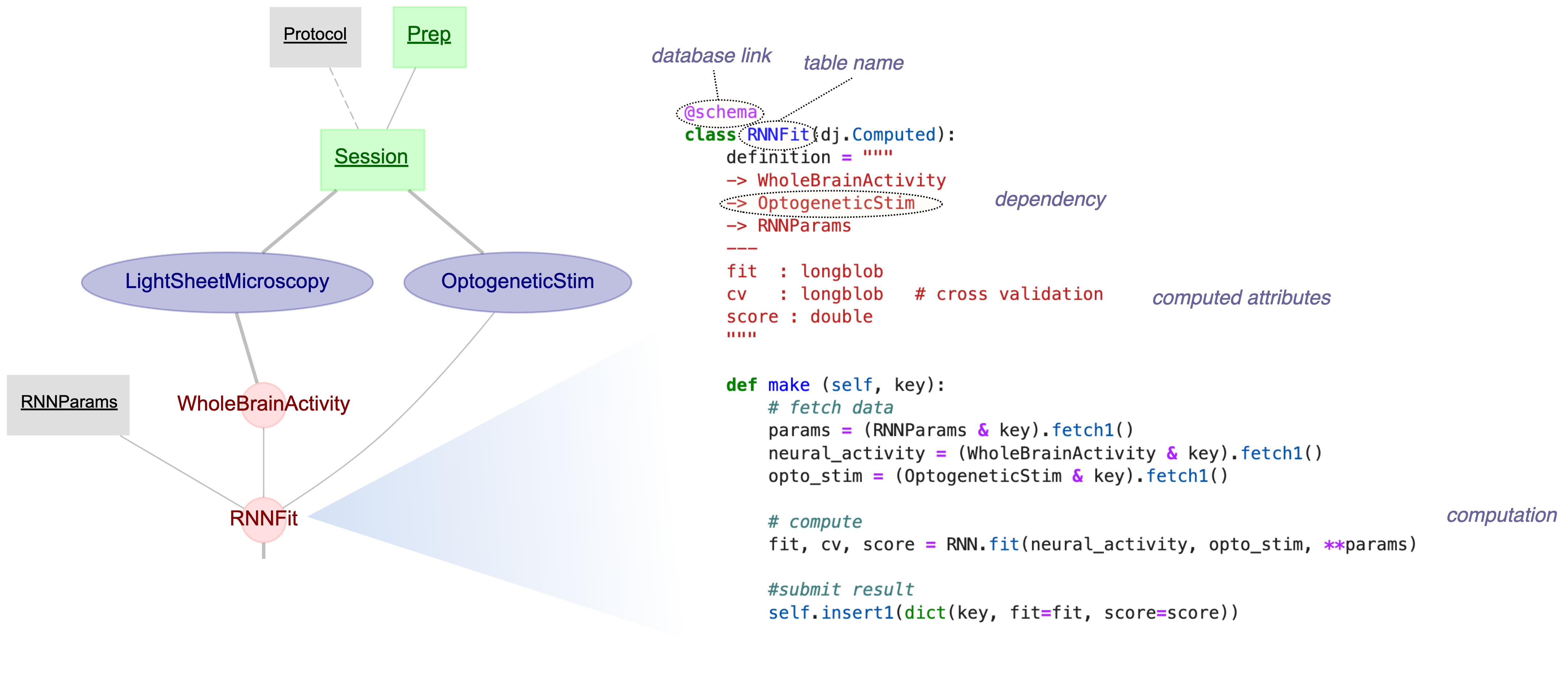
class _RawData(dj.Imported):= """ -> Session --- file_path: varchar(255) data: longblob """ def make(self , key):= load_file(key['file_path' ])self .insert1({** key, 'data' : data})
Computed Tables
class __Processed(dj.Computed):= """ -> _RawData --- processed_data: longblob computed_at: datetime """ def make(self , key):= (_RawData & key).fetch1()= process(raw['data' ])self .insert1({** key, 'processed_data' : processed})
Master-Part Tables
class Session(dj.Manual):= """ -> Subject session_id: int --- session_date: date """ class Trial(dj.Part):= """ -> Session trial_id: int --- trial_start: float trial_end: float """
Complete Pipeline Example
Tables from code examples:
Tables
StimulusType
#StimulusType
[Lookup]
Subject
Subject
[Manual]
Session
Session
[Manual]
Subject->Session
-> Subject
SessionTrial
Session.Trial
[Part]
Session->SessionTrial
Part of
RawData
_RawData
[Imported]
Session->RawData
-> Session
Processed
__Processed
[Computed]
RawData->Processed
-> _RawData
Foreign Keys & Dependencies
Dependencies
Subject
Subject
Session
Session
Subject->Session
_RawData
_RawData
Session->_RawData
__Processed
__Processed
_RawData->__Processed
Query Operations
# Restrict - filter rows matching conditions & {'subject_id' : 1 } # Only sessions for subject 1 # Join - combine tables * Trial# Project - select specific attributes 'subject_name' )
Auto-Population
# Automatically compute missing data # Check progress
Architecture
Architecture
Python\nSchema
Python
Schema
DataJoint\nFramework
DataJoint
Framework
Python\nSchema->DataJoint\nFramework
SQL Database
SQL Database
DataJoint\nFramework->SQL Database
Object Storage
Object Storage
DataJoint\nFramework->Object Storage
Real-World Example: Ephys Pipeline
Element Array Electrophysiology - Neuropixels analysis with Kilosort
EphysPipeline
#Probe
#Probe
EphysSession
EphysSession
#Probe->EphysSession
Subject
Subject
Subject->EphysSession
EphysSession.Channel
EphysSession.Channel
EphysSession->EphysSession.Channel
_RawEphys
_RawEphys
EphysSession->_RawEphys
__SpikeSorting
__SpikeSorting
_RawEphys->__SpikeSorting
__QualityMetrics
__QualityMetrics
__SpikeSorting->__QualityMetrics
Ephys Pipeline Resources
📦 GitHub Repository: https://github.com/datajoint/element-array-ephys
📖 Documentation: https://docs.datajoint.com/elements/element-array-ephys/
Alternatives: Raw SQL
= mysql.connector.connect (...)= conn.cursor()"SELECT * FROM raw_data WHERE processed=0" )= cursor.fetchall()for row in raw_data:= process(row['data' ])"INSERT INTO processed ..." )
Alternatives: SQLAlchemy
# ORM for data access class Session(Base):= 'sessions' id = Column(Integer, primary_key= True )= Column(Integer, ForeignKey('subjects.id' ))
DataJoint Limitations
Limitations:
Schema-driven (less flexible)
Learning curve for new concepts
Not designed for web apps
Domain-specific (scientific focus)